Human Brain Proteoform Atlas
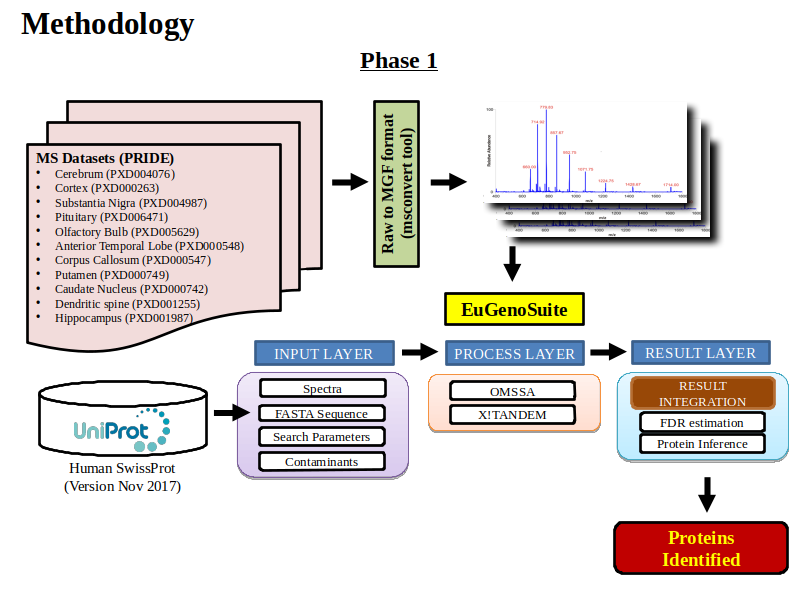
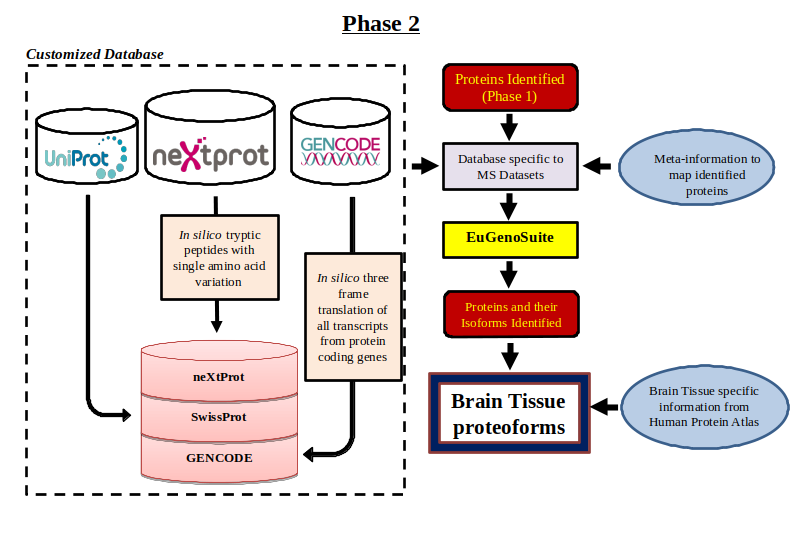
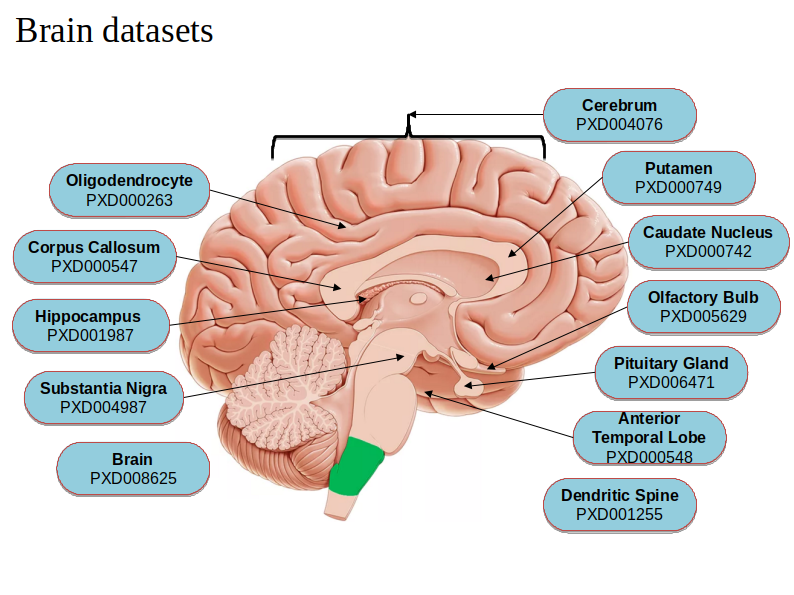
Despite the ongoing refinement of the human proteome, there are still a lot of missing or unexplored regions in the proteome which has sufficient evidence at the transcript level. This gap in the knowledge of proteome can be filled by studying proteoforms that majorly arise from sequence polymorphism, alternative splicing and proteolysis. Exploring proteoforms will provide insights into the fundamental working of biological systems as well as could explain disease inception. Identifying these 6 million estimated proteoforms has always remained challenging as the peptides supporting them are highly homologous which need an efficient protein inference solution. A strategy is devised to explore the human tissue-specific proteoforms which will help to detect proteoform diversity and discover their regulation among different tissues. Publicly available mass spectrometry datasets like PRIDE contains data from various human tissues which will be searched against the in-house-built customized database containing different transcripts and amino acids variant information from GENCODE and neXtProt using a multi-algorithmic proteogenomic approach. Currently, we have analyzed brain tissue-specific datasets from different regions of the brain to identify several known and novel brain proteoforms and developed a repository for the human brain-specific proteoforms.
Our Team




Contact Us
For any queries, please email us at the following address:
Dr. Debasis Dash
ddash@igib.res.in
Or you can mail us at the following address,
CSIR-IGIB,
Mathura Road, Sukhdev Vihar,
New Delhi, Delhi, India - 110025
